In addition to dozens of researchers, the Molecule Maker Lab Institute is supported by a number of facilities at the three Partner Institutions:
Molecule Maker Lab (MML)

Molecule Maker Lab (MML) is a first-of-its-kind facility housed at the Beckman Institute designed to democratize molecular synthesis. The MML currently houses automated synthesis robots for making peptides, oligonucleotides, and small molecules.
The facility will soon also include state-of-the-art LC-MS instruments for automatically characterizing and purifying reaction products (similar instruments in other labs are currently being used).
The facility also houses a collection of computers that operate the synthesis and purification instruments and connect the MML to the cloud.
Automated synthesizers
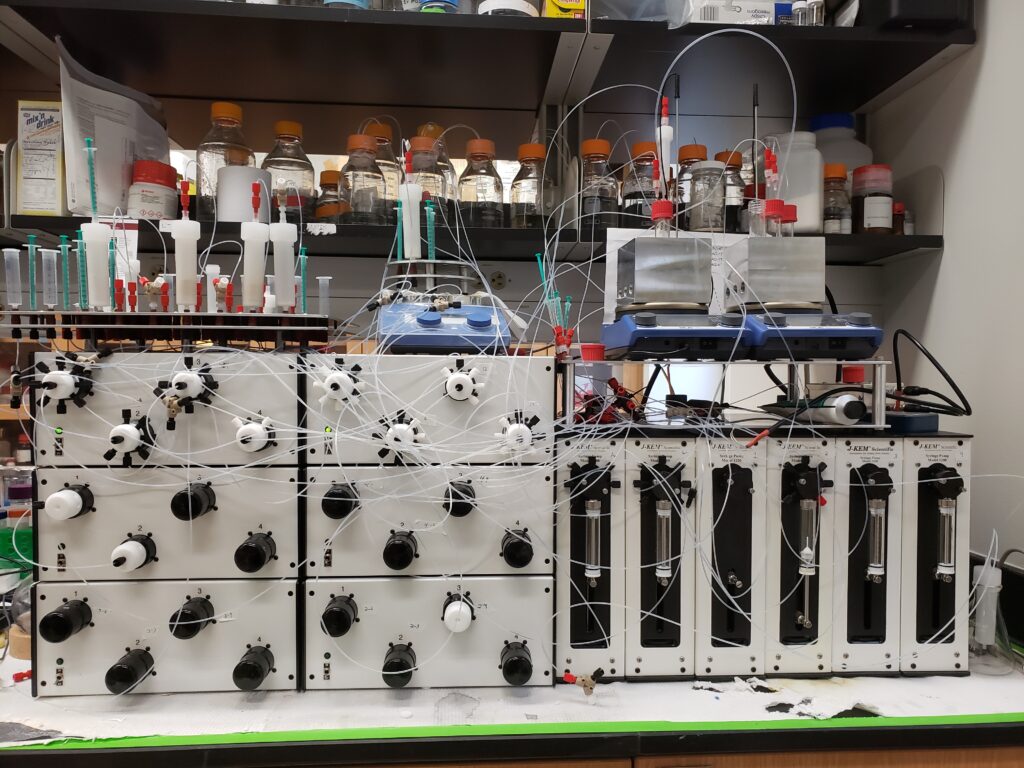
The Burke group created a small molecule synthesizer that enables a wide range of functional small molecules to be made automatically via iterative C—C and/or C—X bond formation.
Recently, the Schroeder group built a second-generation small molecule synthesizer. This machine has enabled the automated synthesis of libraries of sequence-defined, electronically active oligomers at the MML.
Illinois Biofoundry for Advanced Biomanufacturing (iBioFAB)


A fully integrated robotic system called iBioFAB, which is located in the Carl R. Woese Institute for Genomic Biology (IGB), aims to accelerate the biological engineering process by integrating artificial intelligence/machine learning with automation. This iBioFAB consists of component instruments, an integrated robotic platform, and a modular computational framework.
For component instrument selection, routine biological engineering workflows are broken into a series of shared process modules involving different subsets of ten most common unit operations. To manage virtual networks of instrument, iScheduler was developed as a high-level modular programming environment. In addition, a laboratory information management system (LIMS) was installed for sample tracking and strain database management.
In the past few years, iBioFAB was used for rapid engineering of proteins, pathways, and genomes. iBioFAB was recently combined with machine learning algorithms to fully automate the DBTL process for biosystems design (BioAutomata). Learn more
National Center for Supercomputing Applications (NCSA)

NCSA at the University of Illinois has created and maintained many databases, serving as a hub of research, development, and operations activities around scientific cyberinfrastructure spanning disciplines as well as industry.
Tools and components developed as part of activities within the Data Infrastructure Building Blocks (DIBBs), Cyberinfrastructure for Sustained Scientific Innovation (CSSI), and others, provide support for open science as well as capabilities such as data movement, data wrangling, data sharing/discovery/management, data analytics, and scientific gateways, user friendly web interfaces that provide approachable means of accessing high performance resources, distributed storage systems, and sophisticated algorithms.
Such cyberinfrastructure will be used to support MMLI in the collecting, sharing, and AI based mining of chemical reactions along with the development of novel algorithm and models in support of that. Learn more